A Selection of eDNA Publications
Please note this collection is not exhaustive, and there are many eDNA publications world-wide. If you would like to add your own paper or have other publication suggestions, please fill out the Publication Submission form at the bottom of the page.
Publications featuring community authors

Temperature Controls eDNA Persistence across Physicochemical Conditions in Seawater
- (McCartin et al, 2022,
- Environmental Science & Technology)
Environmental DNA (eDNA) quantification and sequencing are emerging techniques for assessing biodiversity in marine ecosystems. Environmental DNA can be transported by ocean currents and may remain at detectable concentrations far from its source depending on how long it persists. Thus, predicting the persistence time of eDNA is crucial to defining the spatial context of the information derived from it. To investigate the physicochemical controls of eDNA persistence, we performed degradation experiments at temperature, pH, and oxygen conditions relevant to the open ocean and the deep sea.

Environmental DNA surveys of African biodiversity: State of knowledge, challenges, and opportunities
- (von der Heyden et al., 2022,
- Environmental DNA)
Environmental DNA surveys have become a well-established tool for detecting natural communities, showing excellent promise for supporting biodiversity monitoring, conservation, and management efforts. Africa is a continent of exceptional biodiversity, threatened not only by anthropogenic pressures but also by a general lack of research capacity and infrastructure, limiting evaluation and monitoring of ecosystems. This commentary explores the use of environmental DNA in surveying natural diversity, a rapidly moving field, within the context of capturing Africa’s natural capital.
Environmental DNA (eDNA): Powerful technique for biodiversity conservation
- (Sahu et al., 2023,
- Journal for Nature Conservation)
Environmental DNA (eDNA) metabarcoding is a non-invasive method for discovering and identifying rare and endangered species in a variety of ecosystems, including aquatic environments, based on the retrieval of genetic traces emitted into the environment by animals. Environmental (e) DNA research has grown in popularity over the last decade as a result of a rise in the number of studies that employ DNA taken from the environment, particularly in freshwater and marine ecosystems. In terms of detecting diversity patterns, we may claim that DNA retrieved from the environment (eDNA) is altering the game. For resource management in fisheries, information on species composition and biomass/abundance of commercially and noncommercially harvested species is critical. The eDNA is a truly non-invasive method that inflicts no damage on the species or habitats under study even during sampling, the eDNA technique never harms any ecosystems or threatened species. This novel molecular method never affects any endangered species or ecosystem during sampling. Environmental DNA analysis has become more widely accepted and is used in the detection of the presence and absence of aquatic macrofauna, such as freshwater and marine fish. This review study may aid researchers in better understanding the current state of eDNA technology. Despite the fact that various scientists have used eDNA to investigate the worldwide biodiversity of aquatic environments, no one in India is focusing on this new technology. We conclude that the eDNA technique has the potential to become a next-generation tool for biodiversity research and aquatic ecosystem conservation.
Publications featuring eDNA Collaborative authors
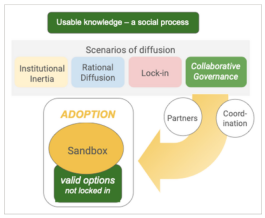
Adoption of environmental DNA in public agency practice
- (Lee et al, 2023,
- Environmental DNA)
Environmental DNA (eDNA) analysis has matured to the point that it is ready for deployment in many applications, particularly in aquatic environments. But public agencies have yet to adopt eDNA methods into their environmental decision making routines at scale, even when eDNA offers clear advantages to those now in use. This article provides a perspective on this gap by considering adoption of a new technology as a path-dependent, social process in which some paths lead to outcomes that provide far greater benefits than others.
Centering accessibility, increasing capacity, and fostering innovation in the development of international eDNA standards
- (Hirsch et al., 2023,
- Metabarcoding and Metagenomics)
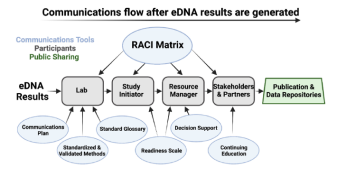
Critical considerations for communicating environmental DNA science
- (Stein et al, 2023,
- Environmental DNA)
The economic and methodological efficiencies of environmental DNA (eDNA) based survey approaches provide an unprecedented opportunity to assess and monitor aquatic environments. However, instances of inadequate communication from the scientific community about confidence levels, knowledge gaps, reliability, and appropriate parameters of eDNA-based methods have hindered their uptake in environmental monitoring programs and, in some cases, has created misperceptions or doubts in the management community. To help remedy this situation, scientists convened a session at the Second National Marine eDNA Workshop to discuss strategies for improving communications with managers.
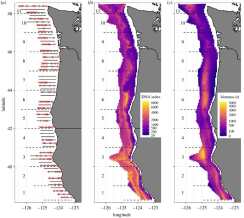
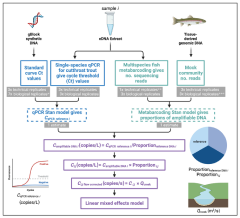
Environmental DNA provides quantitative estimates of Pacific hake abundance and distribution in the open ocean
- (Shelton et al. 2022,
- Proceedings of the Royal Society B, Biological Sciences)
We sampled eDNA in parallel with a traditional acoustic-trawl survey to assess the value of eDNA surveys at a scale relevant to fisheries management. Despite local differences, the two methods yield comparable information about the broad-scale spatial distribution and abundance. Furthermore, we find depth and spatial patterns of eDNA closely correspond to acoustic-trawl estimates for hake. We demonstrate the power and efficacy of eDNA sampling for estimating abundance and distribution and move the analysis of eDNA data beyond sample-to-sample comparisons to management relevant scales. We posit that eDNA methods are capable of providing general quantitative applications that will prove especially valuable in data-or resource-limited contexts.

Improved biodiversity detection using a large-volume environmental DNA sampler with in situ filtration and implications for marine eDNA sampling strategies
- (Govindarajan et al, 2022,
- Deep Sea Research Part I: Oceanographic Research)
Metabarcoding analysis of environmental DNA samples is a promising new tool for marine biodiversity and conservation. Typically, seawater samples are obtained using Niskin bottles and filtered to collect eDNA. However, standard sample volumes are small relative to the scale of the environment, conventional collection strategies are limited, and the filtration process is time consuming. To overcome these limitations, we developed a new large – volume eDNA sampler with in situ filtration, capable of taking up to 12 samples per deployment.
Quantifying impacts of an environmental intervention using environmental DNA
- (Allan et al. 2023,
- Ecological Applications)
Environmental laws around the world require some version of an environmental-impact assessment surrounding construction projects and other discrete instances of human development. Information requirements for these assessments vary by jurisdiction, but nearly all require an analysis of the biological elements of ecosystems. Amplicon-sequencing—also called metabarcoding—of environmental DNA (eDNA) has made it possible to sample and amplify the genetic material of many species present in those environments, providing a tractable, powerful, and increasingly common way of doing environmental-impact analysis for development projects. Here, we analyze an 18-month time series of water samples taken before, during, and after two culvert removals in a salmonid-bearing freshwater stream. We also sampled multiple control streams to develop a robust background expectation against which to evaluate the impact of this discrete environmental intervention in the treatment stream.
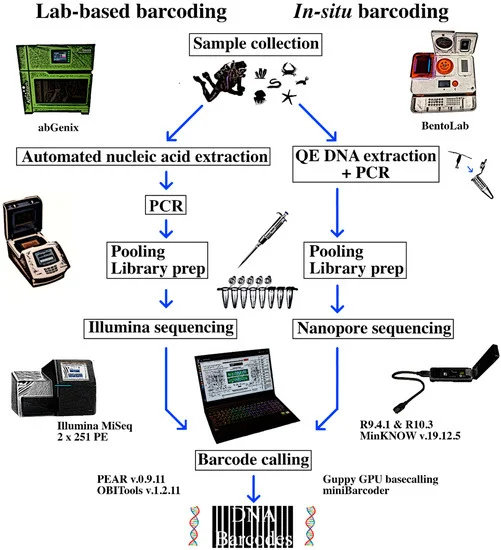
Takeaways from Mobile DNA Barcoding with BentoLab and MinION
- (Chang et al,
- 2020, Genes, Special Issue: MetaGenomics Sequencing In Situ)
Since the release of the MinION sequencer in 2014, it has been applied to great effect in the remotest and harshest of environments, and even in space. One of the most common applications of MinION is for nanopore-based DNA barcoding in situ for species identification and discovery, yet the existing sample capability is limited (n ≤ 10). Here, we assembled a portable sequencing setup comprising the BentoLab and MinION and developed a workflow capable of processing 32 samples simultaneously.
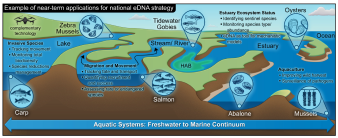
Toward a national eDNA strategy for the United States
- (Kelly et al. 2023,
- Environmental DNA)
Environmental DNA (eDNA) data make it possible to measure and monitor biodiversity at unprecedented resolution and scale. As use-cases multiply and scientific consensus grows regarding the value of eDNA analysis, public agencies have an opportunity to decide how and where eDNA data fit into their mandates. Within the United States, many federal and state agencies are individually using eDNA data in various applications and developing relevant scientific expertise. A national strategy for eDNA implementation would capitalize on recent scientific developments, providing a com- mon set of next-generation tools for natural resource management and public health protection.
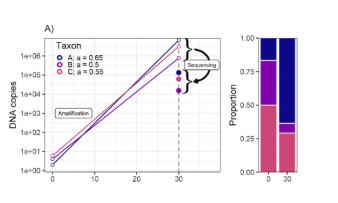
Toward quantitative metabarcoding
- (Shelton et al. 2022,
- Ecological Society of America)
At present, eDNA metabarcoding studies struggle to link genetic observations to underlying biology in a quantitative way, in large part due to biases during the PCR amplification process. Here we link previously disparate sets of techniques for making such data quantitative, showing that the underlying PCR mechanism explains observed patterns of amplicon data in a general way. By modeling the process through which amplicon-sequence data arises, rather than transforming the data post-hoc, we can estimate the starting proportions of DNA for many taxa simultaneously. This model can be calibrated with a variety of methods, including mock communities and variable-PCR-cycle sequencing runs, which we illustrate using simulations and in a series of empirical examples. Our approach opens the door to a wide range of applications in ecology, public health, and related fields.

Tracking an invasion front with environmental DNA
- (Keller et al. 2022,
- Ecological Applications)
We jointly modeled eDNA via qPCR and traditional trap data to estimate the density of invasive European green crab (Carcinus maenas), a species for which, historically, baited traps have been used for both detection and control. Our analytical framework simultaneously quantifies uncertainty in both detection methods and provides a robust way of integrating different data streams into management processes. Moreover, the joint model makes clear the marginal information benefit of adding eDNA data to an existing monitoring program, offering a path to optimizing sampling efforts for species of management interest. Here, we document green crab eDNA beyond the previously known invasion front and find that the value of eDNA dramatically increases with low population densities and low traditional sampling effort, as is often the case at leading-edge locations.
Submit Your Publication
The eDNA Collaborative welcomes publication suggestions, yours or someone else’s, for addition to this page. Please submit your suggestion using the form provided. Be sure to include your reason for the recommendation in the “Justification” field.