Collaborative
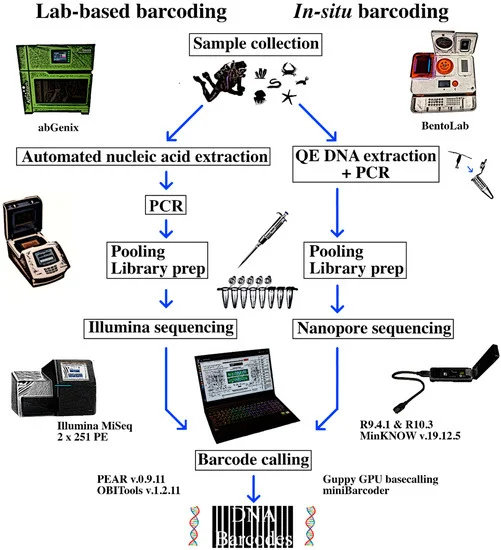
A Barcoding with BentoLab and MinION
Since the release of the MinION sequencer in 2014, it has been applied to great effect in the remotest and
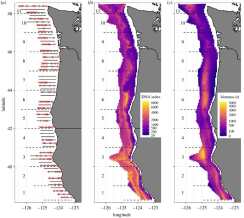
Improved biodiversity detection using a large-volume environmental DNA sampler with in situ filtration and implications for marine eDNA sampling strategies
Metabarcoding analysis of environmental DNA samples is a promising new tool for marine biodiversity and conservation. Typically, seawater samples are
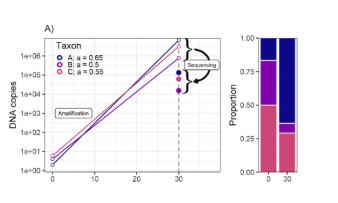
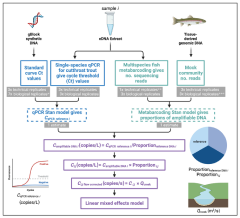
Toward quantitative metabarcoding
At present, eDNA metabarcoding studies struggle to link genetic observations to underlying biology in a quantitative way, in large part
Tracking an invasion front with environmental DNA
We jointly modeled eDNA via qPCR and traditional trap data to estimate the density of invasive European green crab (Carcinus
Environmental DNA provides quantitative estimates of Pacific hake abundance and distribution in the open ocean
We sampled eDNA in parallel with a traditional acoustic-trawl survey to assess the value of eDNA surveys at a scale
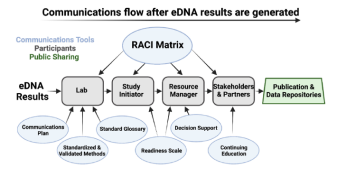
Critical considerations for communicating environmental DNA science
The economic and methodological efficiencies of environmental DNA (eDNA) based survey approaches provide an unprecedented opportunity to assess and monitor
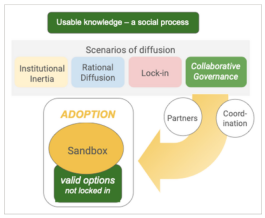
Adoption of environmental DNA in public agency practice
Environmental DNA (eDNA) analysis has matured to the point that it is ready for deployment in many applications, particularly in
Quantifying impacts of an environmental intervention using environmental DNA
Environmental laws around the world require some version of an environmental-impact assessment surrounding construction projects and other discrete instances of
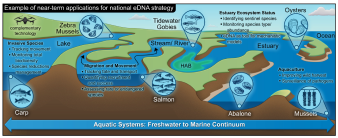
Toward a national eDNA strategy for the United States
Environmental DNA (eDNA) data make it possible to measure and monitor biodiversity at unprecedented resolution and scale. As use-cases multiply